Publications
Here is a list of the group's current publications. It's inevitably out-of-date, but we try our best...
2021 | NATURE COMMUNICATIONS
A method for intuitively extracting macromolecular dynamics from structural disorder
Pearce, Nicholas M; Gros, Piet;
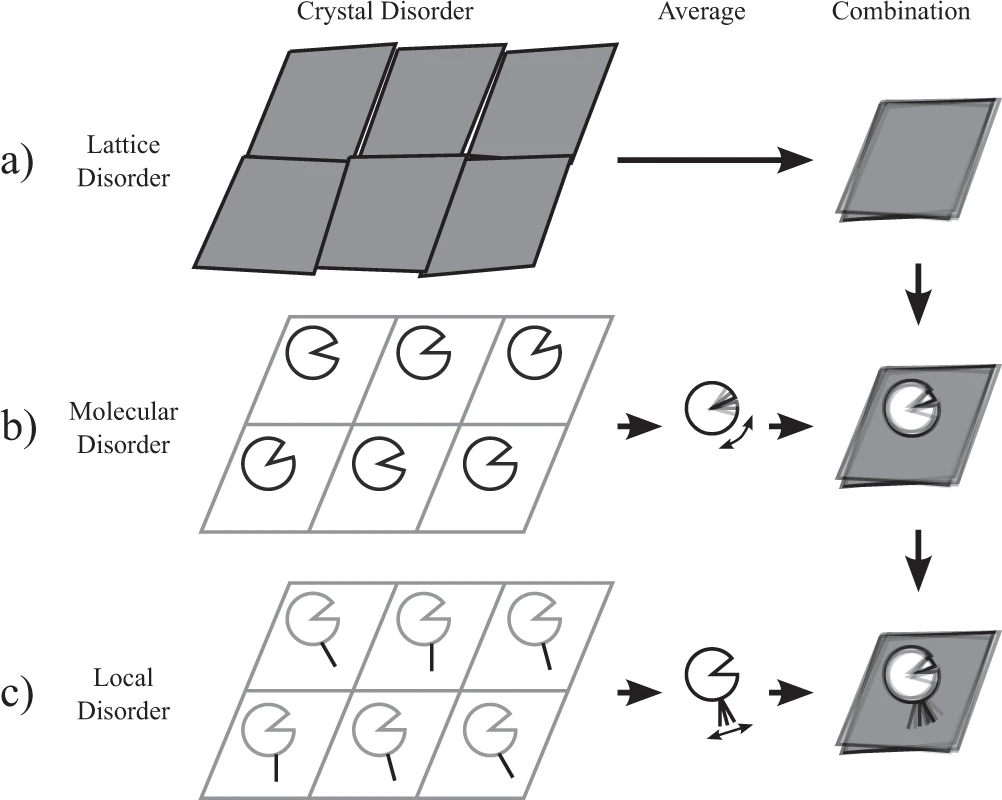
As molecules move in the crystal, this causes a blurring of their electron density, which is captured by B-factors in the atomic model. This disorder therefore captures the macromolecular dynamics happening in the crystal.
We developed the ECHT (Extensible-Component Hierarchical TLS) model, a multi-layered disorder model with components for different scales of motion. By optimising this model with an elastic net approach, we find the parsimonious model for disorder, and thereby reveal likely macromolecular motions and flexbility within the crystal.
2017 | NATURE COMMUNICATIONS
A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density
Pearce, Nicholas M; Krojer, Tobias; Bradley, Anthony R; Collins, Patrick; Nowak, Radosław P; Talon, Romain; Marsden, Brian D; Kelm, Sebastian; Shi, Jiye; Deane, Charlotte M;
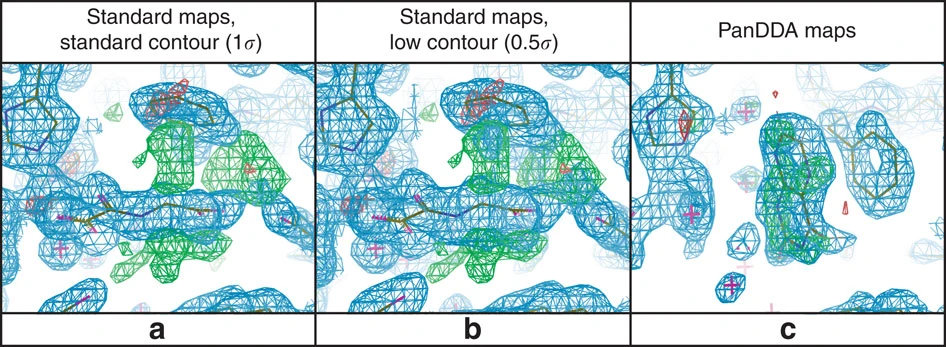
The PanDDA (Pan-Dataset Density Analysis) method identifies partial-occupancy features in crystallographic data, such as weakly-binding ligands. These appear at sub-unitary occupancy in the crystal and are thus hard to detect and model.
We developed an approach where we build a statistical model of a set of crystallographic datasets and finds binding ligands by finding electron density outliers from the population. We then apply a background correction to account for the partial occupancy of the ligand and present density for the ligand so that it can be modelled.
2022 | NAN
Crystallography Made Crystal Clear: A Guide for Users of Macromolecular Models
Pearson, Arwen R; Pearce, Nicholas; Wierman, Jennifer;
| Date | Journal | Title | Authors |
|---|---|---|---|
| 2022 | Chemical science |
Mechanistic insights into the C 55-P targeting lipopeptide antibiotics revealed by structure–activity studies and high-resolution crystal structures |
Wood, Thomas M; Zeronian, Matthieu R; Buijs, Ned; Bertheussen, Kristine; Abedian, Hanieh K; Johnson, Aidan V; Pearce, Nicholas M; Lutz, Martin; Kemmink, Johan; Seirsma, Tjalling; |
| 2022 | nan |
Crystallography Made Crystal Clear: A Guide for Users of Macromolecular Models |
Pearson, Arwen R; Pearce, Nicholas; Wierman, Jennifer; |
| 2022 | Frontiers in Molecular Biosciences |
Experiences from developing software for large X-ray crystallography-driven protein-ligand studies |
Krojer, Tobias; Pearce, Nicholas Mark; Skyner, Rachael; |
| 2021 | Angewandte Chemie |
Bicyclic β‐Sheet Mimetics that Target the Transcriptional Coactivator β‐Catenin and Inhibit Wnt Signaling |
Wendt, Mathias; Bellavita, Rosa; Gerber, Alan; Efrém, Nina‐Louisa; van Ramshorst, Thirza; Pearce, Nicholas M; Davey, Paul RJ; Everard, Isabel; Vazquez‐Chantada, Mercedes; Chiarparin, Elisabetta; |
| 2021 | Acta Crystallographica Section D: Structural Biology |
Improving sampling of crystallographic disorder in ensemble refinement |
Ploscariu, Nicoleta; Burnley, Tom; Gros, Piet; Pearce, Nicholas M; |
| 2021 | Nature communications |
A method for intuitively extracting macromolecular dynamics from structural disorder |
Pearce, Nicholas M; Gros, Piet; |
| 2021 | Biochemical Journal |
Crystallographic approach to fragment-based hit discovery against Schistosoma mansoni purine nucleoside phosphorylase |
Faheem, Muhammad; Fonseca Valadares, Napoleão; Brandão-Neto, José; Bellini, Dom; Collins, Patrick; Pearce, Nicholas M; Bird, Louise; Roberta Torini, Juliana; Owens, Raymond; DMuniz Pereira, Humberto; |
| 2021 | JoVE (Journal of Visualized Experiments) |
Achieving efficient fragment screening at XChem facility at diamond light source |
Douangamath, Alice; Powell, Ailsa; Fearon, Daren; Collins, Patrick M; Talon, Romain; Krojer, Tobias; Skyner, Rachael; Brandao-Neto, Jose; Dunnett, Louise; Dias, Alexandre; |
| 2020 | Life science alliance |
Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F |
Oosterheert, Wout; Xenaki, Katerina T; Neviani, Viviana; Pos, Wouter; Doulkeridou, Sofia; Manshande, Jip; Pearce, Nicholas M; Kroon-Batenburg, Loes MJ; Lutz, Martin; en Henegouwen, Paul MP van Bergen; |
| 2019 | Frontiers in Immunology |
Insights into enhanced complement activation by structures of properdin and its complex with the C-terminal domain of C3b |
Van den Bos, Ramon M; Pearce, Nicholas M; Granneman, Joke; Brondijk, T Harma C; Gros, Piet; |
| 2018 | Elife |
An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering |
Keedy, Daniel A; Hill, Zachary B; Biel, Justin T; Kang, Emily; Rettenmaier, T Justin; Brandao-Neto, Jose; Pearce, Nicholas M; von Delft, Frank; Wells, James A; Fraser, James S; |
| 2017 | Structural Dynamics |
Partial-occupancy binders identified by the Pan-Dataset Density Analysis method offer new chemical opportunities and reveal cryptic binding sites |
Pearce, Nicholas M; Bradley, Anthony R; Krojer, Tobias; Marsden, Brian D; Deane, Charlotte M; Von Delft, Frank; |
| 2017 | Acta Crystallographica Section D: Structural Biology |
Proper modelling of ligand binding requires an ensemble of bound and unbound states |
Pearce, Nicholas M; Krojer, Tobias; Von Delft, Frank; |
| 2017 | Nature communications |
A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density |
Pearce, Nicholas M; Krojer, Tobias; Bradley, Anthony R; Collins, Patrick; Nowak, Radosław P; Talon, Romain; Marsden, Brian D; Kelm, Sebastian; Shi, Jiye; Deane, Charlotte M; |
| 2017 | Acta Crystallographica Section D: Structural Biology |
The XChemExplorer graphical workflow tool for routine or large-scale protein–ligand structure determination |
Krojer, Tobias; Talon, Romain; Pearce, Nicholas; Collins, Patrick; Douangamath, Alice; Brandao-Neto, Jose; Dias, Alexandre; Marsden, Brian; Von Delft, Frank; |
| 2017 | PLOS Computational Biology |
Ten simple rules for surviving an interdisciplinary PhD |
Demharter, Samuel; Pearce, Nicholas; Beattie, Kylie; Frost, Isabel; Leem, Jinwoo; Martin, Alistair; Oppenheimer, Robert; Regep, Cristian; Rukat, Tammo; Skates, Alexander; |
| 2017 | Acta Crystallographica Section D: Structural Biology |
Gentle, fast and effective crystal soaking by acoustic dispensing |
Collins, Patrick M; Ng, Jia Tsing; Talon, Romain; Nekrosiute, Karolina; Krojer, Tobias; Douangamath, Alice; Brandao-Neto, Jose; Wright, Nathan; Pearce, Nicholas M; Von Delft, Frank; |