PanDDA
Publications
Highlighted publications
2017 | NATURE COMMUNICATIONS
A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density
Pearce, Nicholas M; Krojer, Tobias; Bradley, Anthony R; Collins, Patrick; Nowak, Radosław P; Talon, Romain; Marsden, Brian D; Kelm, Sebastian; Shi, Jiye; Deane, Charlotte M;
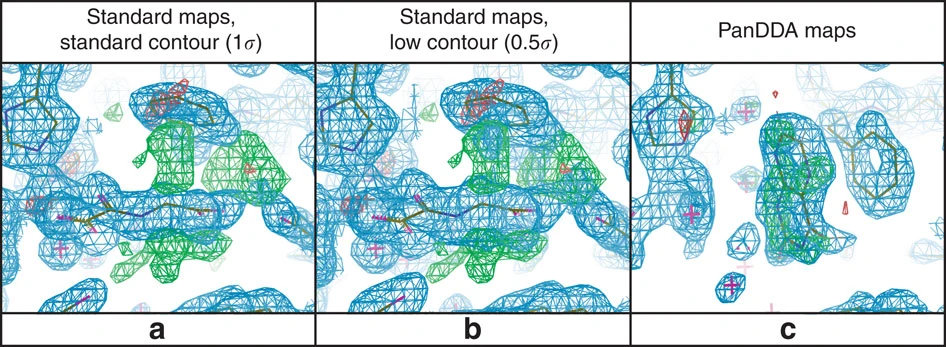
The PanDDA (Pan-Dataset Density Analysis) method identifies partial-occupancy features in crystallographic data, such as weakly-binding ligands. These appear at sub-unitary occupancy in the crystal and are thus hard to detect and model.
We developed an approach where we build a statistical model of a set of crystallographic datasets and finds binding ligands by finding electron density outliers from the population. We then apply a background correction to account for the partial occupancy of the ligand and present density for the ligand so that it can be modelled.
All publications
2021 | JOVE (JOURNAL OF VISUALIZED EXPERIMENTS)
Achieving efficient fragment screening at XChem facility at diamond light source
Douangamath, Alice; Powell, Ailsa; Fearon, Daren; Collins, Patrick M; Talon, Romain; Krojer, Tobias; Skyner, Rachael; Brandao-Neto, Jose; Dunnett, Louise; Dias, Alexandre;
2018 | ELIFE
An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering
Keedy, Daniel A; Hill, Zachary B; Biel, Justin T; Kang, Emily; Rettenmaier, T Justin; Brandao-Neto, Jose; Pearce, Nicholas M; von Delft, Frank; Wells, James A; Fraser, James S;
2017 | NATURE COMMUNICATIONS
A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density
Pearce, Nicholas M; Krojer, Tobias; Bradley, Anthony R; Collins, Patrick; Nowak, Radosław P; Talon, Romain; Marsden, Brian D; Kelm, Sebastian; Shi, Jiye; Deane, Charlotte M;
2017 | ACTA CRYSTALLOGRAPHICA SECTION D: STRUCTURAL BIOLOGY
The XChemExplorer graphical workflow tool for routine or large-scale protein–ligand structure determination
Krojer, Tobias; Talon, Romain; Pearce, Nicholas; Collins, Patrick; Douangamath, Alice; Brandao-Neto, Jose; Dias, Alexandre; Marsden, Brian; Von Delft, Frank;
2017 | ACTA CRYSTALLOGRAPHICA SECTION D: STRUCTURAL BIOLOGY
Gentle, fast and effective crystal soaking by acoustic dispensing
Collins, Patrick M; Ng, Jia Tsing; Talon, Romain; Nekrosiute, Karolina; Krojer, Tobias; Douangamath, Alice; Brandao-Neto, Jose; Wright, Nathan; Pearce, Nicholas M; Von Delft, Frank;